
This is a blog version of my presentation for the GoGLAM miniconf at Linux.conf.au 2021 on Arkisto, a framework for describing and preserving research data assets with open source tools and standards.
I converted it from PowerPoint to Markdown with Peter Sefton's pptx_to_md tool (but had to bash it a bit with a hammer to get it to work properly)
The links:
- Arkisto
- OCFL - Oxford Common File Layout
- RO-Crate - Research Object Crates
- Describo
- Modern PARADISEC
- Oni - an index and discovery web app

Hi, and thanks for coming. I’m Mike Lynch and I work in the eResearch Support Group at the University of Technology Sydney. “eResearch”, if you haven’t heard the term before, is a blanket term for the sorts of specialised IT support which academic researchers need. It covers everything from very techy stuff like high-performance computing, to providing advice to researchers and the ethics committee on best practices to protect personal information in health data.
I’m going to talk about some work we’ve been doing to try to make research data description, management and preservation more sustainable. But I’ll start with a quick summary of the problems we’re trying to solve.

Researchers produce a lot of data. What “a lot” means varies a bit between disciplines: traditionally there’s been a habit of seeing this as a STEM problem, but humanities disciplines have big datasets too. Researchers don’t want to describe their data. They want to use it and get publications out of it. We - the eResearch people and librarians - like to help them with this. But there aren’t enough of us to go round. So: we end up with petabytes of storage with fileshares belonging to researchers, full of data, without much metadata — who the data was made by, which grants funded it, or detailed stuff like which microscope generated it, what dye was used for it. This is a problem for sustainability because we can’t make decisions like: do we need to keep this? How long for? Can we move it to slower, cheaper storage?
One of the ways institutions have tried to solve this problem in the past is specialised repository software - either belonging to a discipline, like OMERO, or general, like Fedora (not that Fedora) or Hydra.
Repositories can do things like automatically add metadata when they ingest things, or prompt the researchers to describe them.
They let you put stuff in and get it out with APIs

But repositories have problems. They’re software: often they’re free or open-source, but they’re not free to maintain. And the specialised ones in particular are often built by academics, not software engineers, so they have Problems. People argue about the exact definition of Big Data, I’ve decided that Big Data is data that can make your sysadmin swear, and repositories are good at that. And there will always be a scale issue: at some stage, someone is going to want to put something through the API which just won’t fit. At that point, one solution is sideloading: put the big stuff on a file system with some sort of link between it and the repository software. And now you have to keep them in sync.
And data migration is always a hassle. Arkisto builds on a couple of standards which have taken the lessons of the last decade of repository software development to heart. We already have a stable, performant way of storing data, with decades of history behind it and a promising future:
Files.
A few years ago, we were talking to some colleagues in big data visualisation, who’d also had experience doing CGI for feature films, about managing their data. And asked them if they had a preferred API for accessing data sets.
And they said: “An NFS mount, thanks”
If, at some point, we’re going to have to sideload the big stuff, why not just store everything on disk? But - then - aren’t we back where we started, with our research data store being just a bunch of stuff?

A second problem which Arkisto is trying to help solve is the problem of research data collections. These are different from the institutional research data store: they are single-purpose, and a researcher or team of researchers has put time, love and dedication into curating a dataset, they’ve found a postgrad with some tech skills who’s made a nice website with a LAMP stack, or one of the popular tools like Omeka or Mukurtu, and got their research online.
The grant ends, one or two of the researchers leave, the person who built the site gets a postdoc. And, eventually, a sysadmin is doing an audit of what needs patching, and says - what the hell is this? Will anyone yell if I turn it off?
The scenario I’ve just described happens in science as well as humanities disciplines, though often the scientists use databases for performance reasons.
This sort of bespoke, research-driven collection will always be around, but Arkisto wants to provide more sustainable ways to build them and/or preserve them

We’ve picked the term “Arkisto” to cover the philosophy, standards and tooling we’ve started using to tackle these problems. It’s Finnish for “archive”, and we couldn’t find anyone else who was using it. We decided that the whole thing needed a name when applying for an ARDC grant last year, which we didn’t get, but having a term for it is useful.
Arkisto is a platform for sustainable research data preservation. It’s not a single piece of software, but an approach to building tools and systems based on
- Standards for repository design and metadata storage
- A development philosophy for building tooling and systems

Before diving into the standards, I’ll describe the philosophy. First: data and metadata on disk (or in an object store) is the source of truth, and (where possible) is immutable. You can add to it, but not overwrite it.
Arkisto repositories can have secondary data products which make things findable and more efficient - indexes and so on - but these should be seen as ephemeral and reconstituting them from the data on disk should be easy. Think of this as a devops approach to secondary data products, with declarative config to say how to build them, and automated processes to spin them up.
We don’t want to build monoliths which do everything. Build components which do one thing, like ingesting a collection, or indexing a repository.
Development is incremental, and based on producing changes which in some way make our lives or our user’s lives easier.
A lot of our tooling is in its early stages, and none of it is expected to scale well. We can improve the tooling as we go without having to change the underlying data formats.

We have two main standards. Research Object Crates, or RO-Crates, provide a way to describe each collection in a repository.
The Oxford Common File Layout, OCFL, describes a repository: how we take a set of collections and lay them out on disk. OCFL can be thought of as “outside” and RO-Crate as “inside” each collection.
What constitutes a “collection” depends on the application. It could be a single data publication, or it could be terabytes of data representing years of research.
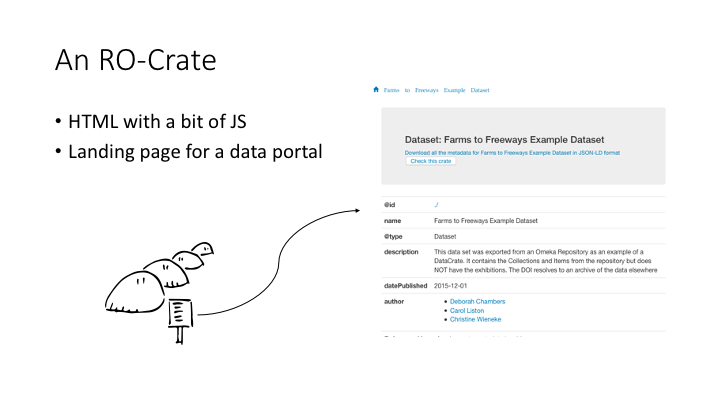
I’ll show you an Arkisto repository from the inside out.
This is what an RO-Crate “looks like” - it’s a publication which we crosswalks from an existing Omeka repository. This is its human-readable face: it’s an HTML landing page with a bit of JavaScript. There’s also a link to the same metadata in a machine readable format. This is a JSON-LD file.
RO-Crates’ are the descendants of two earlier standards, DataCrate and Research Objects, from my colleague and former boss Peter Sefton.
The idea behind RO-Crate is that a dataset should be self-describing after it’s been downloaded. If you unzip the Crate, there’s a decent-looking HTML page you can view in a browser, and the same metadata is there in a machine-readable format.
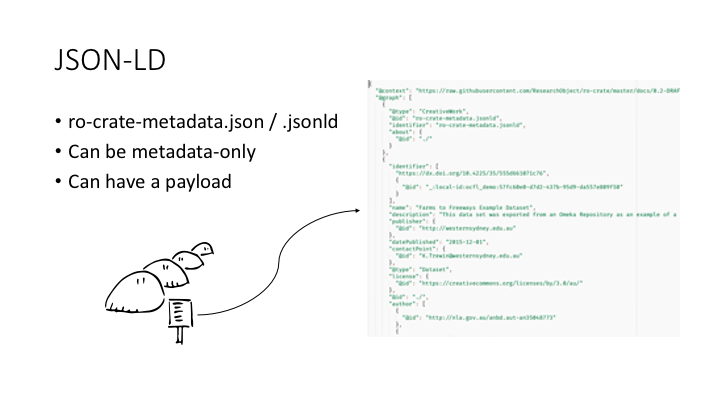
The HTML page I showed you earlier is generated from this JSON-LD document. An RO-Crate is a directory with an rocrate-metadata.json file in it, the HTML landing page, and possibly some payload files, stored in any hierarchy of directories.
It’s perfectly legit for an RO-Crate to have just the metadata files, and sometimes useful: this lets us publish datasets where the payload can’t be made available. Sometimes researchers want to be asked politely before they grant people access to the actual data. Sometimes, they aren’t allowed to share the data at all, but want to be able to share the metadata.
There’s no requirement that every file in the payload have an entry in the RO-Crate. Some datasets contain thousands of files, some contain one or two. The appropriate level of detail is up to the application.
An important feature is that we can turn any directory with stuff in it into an RO-crate by writing the metadata file into the root.
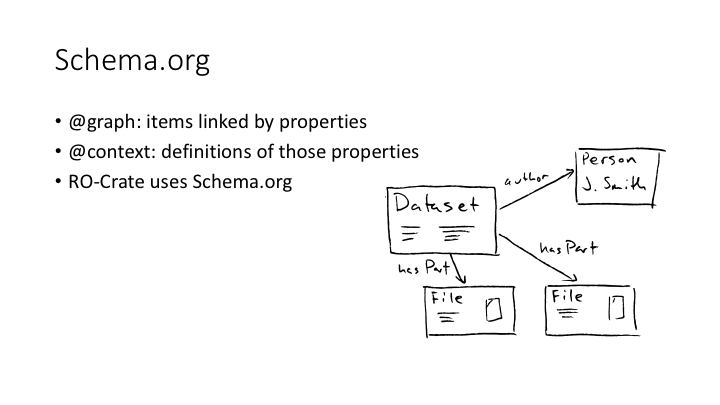
A JSON-LD file has two sections: the @context and the @graph.
The @graph is a list of items, related by properties like “author”, “hasPart”, etc. The @context is a set of URLs which define those properties.
The RO-Crate community decided to use Schema.org as the basic vocabulary for describing datasets. Schema.org had a bit of a bad reputation in scholarly circles for a while: it’s very much the creation of big tech, and has a lot of ways to describe selling things to people and not a lot of detail for scholarly publication. But it’s well-known, and things like Google’s data publications search use it, so RO-Crates on the web get harvested properly.
On the RO-Crate website, there’s a lot of detail about the best ways to use JSON-LD to model typical research metadata, like data provenance — what tools or software produced a file — or data licensing — who is allowed to view and/or reuse this data.
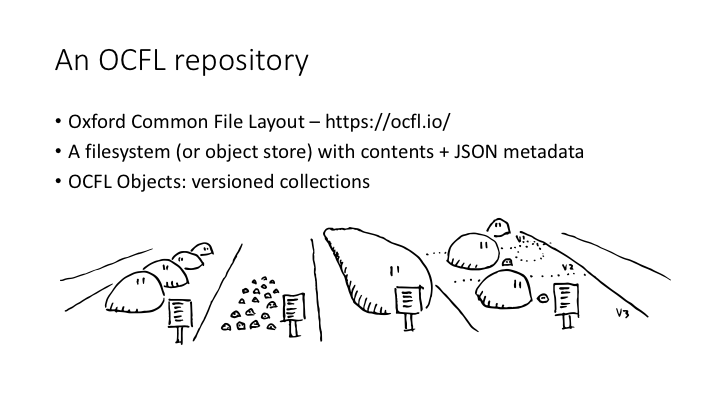
So that’s an RO-Crate. An Arkisto repository is one or more RO-Crates stored as an OCFL repository. OCFL repositories are less fun to look at than RO-Crates, because there’s no nice HTML representation.
OCFL is an acronym for Oxford Common File Layout, although an OCFL repository can use an object store such as s3 for the storage layer.
Like RO-Crates, OCFL repositories are just a filesystem with some contents, and some JSON metadata files about the contents.
The OCFL spec tells us how to do two things:
- How to lay out one or more OCFL Objects on disk. The most common option is a pair-tree algorithm - take an identifier and chop it into two-character chunks and use those as paths.
- how to keep versioned collections inside the OCFL Objects.

The metadata files in an OCFL object are called inventory.json files.
A quick explanation is that an inventory contains two lists. One is a list of a SHA512 hash of every file that’s ever been put into this object, with a path to where it is on the disk.
The second list is a list of versions. Each version has a timestamp, and a list of the hashes of the files which exist in that version. So the entire history of the object’s revisions can be reconstructed.
It’s not designed to be efficient: these versions aren’t to be used like git commits. OCFL shows its origins in the archiving / preservationist world here.
Rather, it’s meant to be relatively easy for someone in the future to inspect this stuff and write their own code to navigate it.

Ok: after that big info-dump, I’ll pose the question: who is all this for? I said at the start that researchers don’t want to describe their data.
Researchers are an audience for RO-Crates, inasmuch as they want the landing pages to look decent when we publish their data. But we can’t expect them to learn JSON-LD.
Likewise, a researcher might like the idea of having a searchable website with their data stored in it, but isn’t going to care about OCFL.
This means that generating RO-Crates and moving things in and out of OCFL repositories needs to be automatic and seamless: the end-users shouldn’t have to worry about it.
Our production systems have been built up incrementally. The first release of our data publication system used DataCrates for their landing pages, one of the immediate ancestors of RO-Crates, and wrote them out to a web server. A subsequent release of the data publications server put the RO-Crates into a pair of OCFL repositories (one for staging, and one for published datasets). Last year, we added a search and discovery portal.
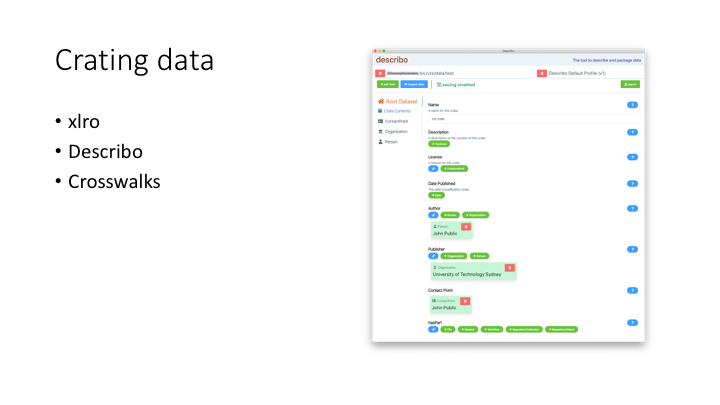
So the hard part of making an RO-Crate is generating the JSON-LD file. We’ve got three ways to do this.
xlro - generates a JSON-LD file by traversing the filesystem and/or reading a spreadsheet
As coders, we are meant to look down our noses at Excel; as wranglers of research data, we can’t do that, because Excel is how the world handles tabular data
describo - an Electron app which looks at a directory and allows you to build an RO-Crate describing the contents. The first version of this looked at local storage. We’re getting it working with OneDrive and hopefully looking at rclone to allow it to connect to cloud-whatever. In its current form, it’s the sort of thing a power user could negotiate, and needs a few iterations of user testing.
crosswalking - the majority of our RO-Crates in production are generated by our research data management system, Stash. This has a web interface for researchers to describe data publications, and uses a simple js library to crosswalk this information into an RO-Crate

Our OCFL toolkit isn’t as mature as our RO-Crate toolkit. Our publications repositories uses the Node ocfl.js library to deposit new publications.
We have CLI tools which use the same library to enable us to deposit things manually into the publications repository: we use these for cases which are too big for the web form
Ingesting is an area where we could improve how our library works. Our current tool is minimal and old-school. It doesn’t give you a process indicator, but just sits there till it’s finished, and if it doesn’t say anything, that means it worked. Some ideas for improvement are progress indicators and caching hashes to improve efficiency.

Oni is a user-facing web application which allows faceted search and discovery of an Arkisto repository.
The name Oni was originally an acronym for OCFL / Nginx / Index. An Oni is a magical creature from Japanese mythology, and I’d seen two impressive monumental sculptures of a Red Oni and a Blue Oni at the Art Gallery of NSW when I was trying to think of a name for it. Oni are slightly shaggy but (in some stories, at least) not as scary as their reputation.
The first version used nginx but the free version wouldn’t do JWT authentication, so we switched to Express.js, a simple Node application server.
So now the ’N’ stands for ‘node’. This is an example of the idea that if you’ve got a good standard to code against, it makes it easy to swap out parts of the tech stack.
Oni scans an OCFL repository, turns the RO-Crate metadata into a Solr index, and provides a discovery interface to that index.

Here’s a screen shot of the front page of the UTS data publications portal: this shows a basic faceted search. It’s a single-page app. Everything you see here has come from the Solr index.
The search results here correspond to the top-level Dataset item in each publication’s RO-Crate. The facets are generated according to the indexer config — so, for example, the configuration for indexing a Dataset says that the “author” property should be a multi-valued facet. Solr then takes care of the faceted search.
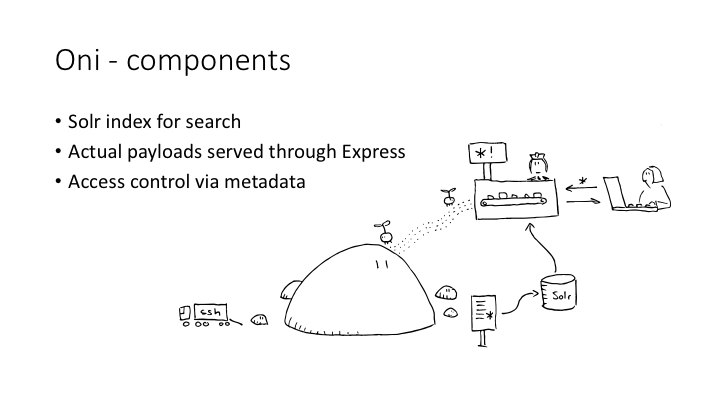
An Oni has three main components: the OCFL repository, the index, and a single-page web app which returns faceted search results.
The HTML landing page and payload files are passed through the Express web app.
We have some fairly experimental code which looks for a licence in the RO-Crate and uses it to authorise whether a user can see that item, both in search results and in the direct web view. This will be a requirement for using Oni to search an internal institutional repository, because we need to be able to control who sees what.
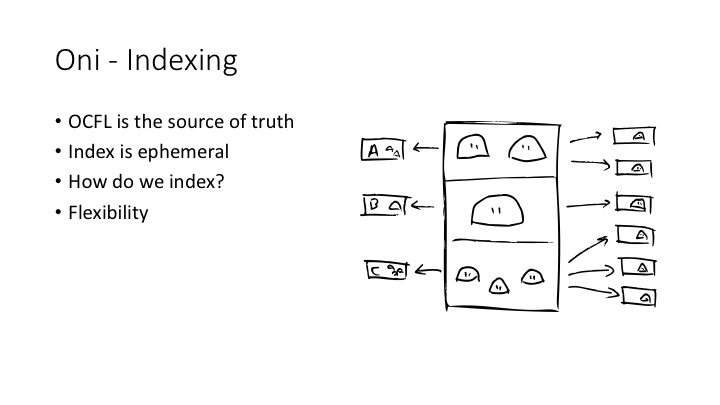
The most important idea behind Oni is that the primary source of truth is the OCFL repo, and the index is ephemeral. It’s generated from the RO-Crates in the repo by a fairly simple, declarative configuration file. A common problem with bespoke data management for research collections is that you need to make decisions about granularity: like, for example, if we’ve got a collection with many datasets, each of which contains hundreds of files, should the search results be at the level of datasets? Or the level of files? This sort of decision can get baked in to both the collection and the database, and changing it later on requires a serious effort.
It also doesn’t make it easy to provide for changes in the model which arise from the research itself. Research data is not corporate data: it’s exploratory by nature, and when applying Oni to datasets from a single project, we want to encourage the researchers to feel free to go where the research takes them, not where our search design says they should.
We also want Oni to be able to index things at different scales: from institutional repositories where the appropriate search result is a single dataset, to large datasets where the search results are entities within that dataset, like a Person or a Place.
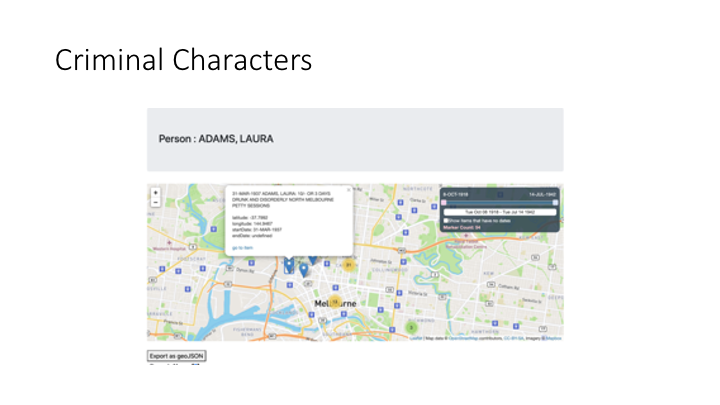
Here’s an example of this . This is a collection from a history research project by Dr Alanah Piper called Criminal Characters — part of her research methodology is to take court records from the 19th century and, using Zooniverse, turn them into linked data structures. So a Person in this collection is linked to a set of Convictions, each of which has a date and a Location — the court where they were convicted. This is a screenshot from some work Peter Sefton’s done on rendering the results in a map viewer, showing how the toolkit we’re building allows search results to be more than just text and links.
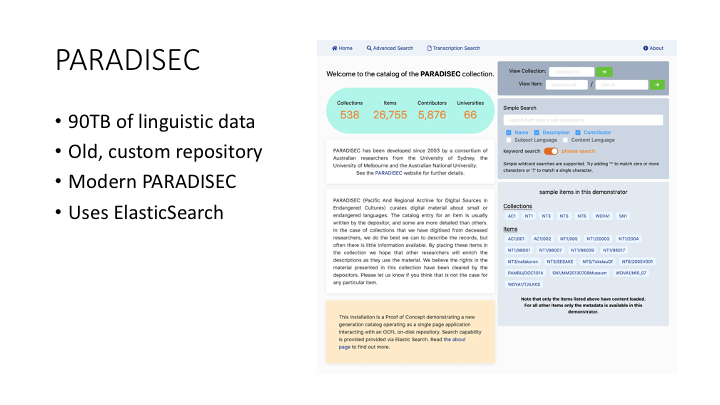
PARADISEC is the Pacific and Regional Archive for Digital Sources in Endangered Cultures, and has acted as a data curation and preservation service for over 90TB of data, mostly in field linguistics - sound recordings, detailed transcripts, videos, and so on. The current repository has been developed over 18 years and is in need of renewal.
In 2020, Marco De La Rosa and Nick Thieberger received a grant from the ARDC to modernise the PARADISEC catalogue using the core Arkisto standards, OCFL and RO-Crate. This is a screen shot of the demonstrator which Marco developed. Note that this is a different web stack: the index was build with ElasticSearch and there’s a custom front-end - this illustrates that the Arkisto platform isn’t meant to restrict choices about programming language, framework or other components.

In 2021 we’re looking at applying the Arkisto principles to the other end of the research data pipeline, using a dedicated OCFL repository to deposit raw CSV data from remote sensors as early as possible, giving us a “live archive”. We can use an Oni to provide access to this data faceted by location and time.
We are looking at extending Oni so that it can do a bit more with the contents of data files — indexing on the column level, not the file — and become, in a way, a data API server. This is all very hand-wavy at the moment. Another way we want to extend Oni as a data server is to look at more useful ways of letting people fetch data. Zipfiles are the way we have been doing it, but they are impractical for a bunch of reasons: you can end up with twice the storage requirements, you don’t really want to keep them in the OCFL repository, and generating them for big datasets is time-consuming.
A better approach might be a feature on Oni which provides the user with a URL from which they can fetch a current result, either using curl or wget, or by porting the data directly into a coding environment like a Jupyter notebook.
And, hopefully, we’ll find more people who are interested in joining us as collaborators.

- Arkisto
- OCFL - Oxford Common File Layout
- RO-Crate - Research Object Crates
- Describo
- Modern PARADISEC
- Oni - an index and discovery web app
Thanks to Dr Peter Sefton, Dr Alana Piper and Dr Marco La Rosa
Michael.Lynch@uts.edu.au / @mikelynch@aus.social